Rationale
Cytoscape is a successfully developed software platform for the visualization and analysis of the biological networks. As one of its coolest features, it provides lots of functional plug-ins and allows users to develop plug-ins in the context of their own work. However, most of the available plug-ins are designed for network analysis, those for network reconstruction are rare yet, let alone those which can reconstruct metabolic networks for the given organism or on the given level by use of the KEGG data directly.
To meet this requirement, we developed MetAtlas, the first Cytoscape plug-in, to reconstruct metabolic networks interactively in the form of WYSIWYG (what you see is what you get). MetAtlas contributes to the direct computational network analysis by working together with the other plug-ins.
Technology
MetAtlas retrieves the metabolic data from the KEGG ftp site using the KEGG API and store data locally using a relational database server. MetAtlas creates graph objects using GINY. MetAtlas can handle the metabolic network reconstruction in two kinds of representations (enzyme graph) and on three levels in line with the level defined in the KEGG Orthology.
MetAtlas is developed with Eclipse, the Java Development Kit (JDK) 1.5 and Cytoscape Plug-in API. The use of Spring and Maven makes the source code of MetaGen quite stretchable and seamless. Using the standard SQL syntax, MetaGen ships pre-configured with the MySQL database. But one can easily switch to the database they are comfortable administering by making a little change in the table schema SQL file. MetAtlas needs nothing to run but JDK5.0 or higher, Cytoscape 2.6.x and a rational database which supports JDBC, but MetAtlas is platform-independent and database-independent. User can follow the documents in the downloaded package to install MetAtlas and make it work.
Screenshots
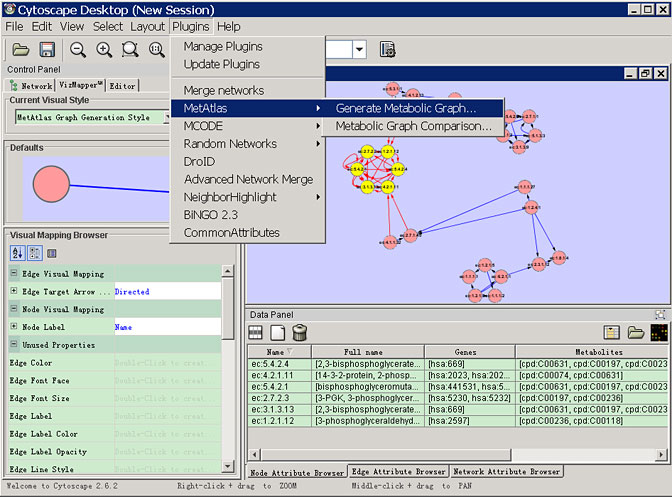
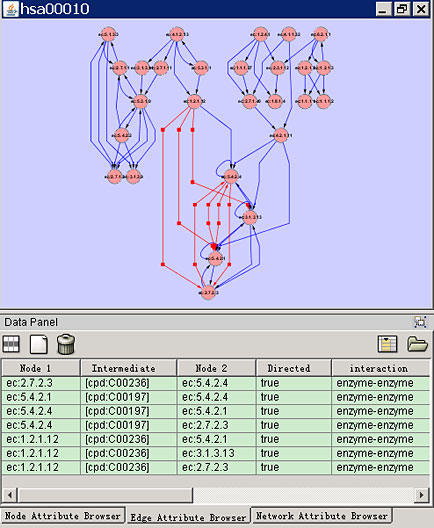
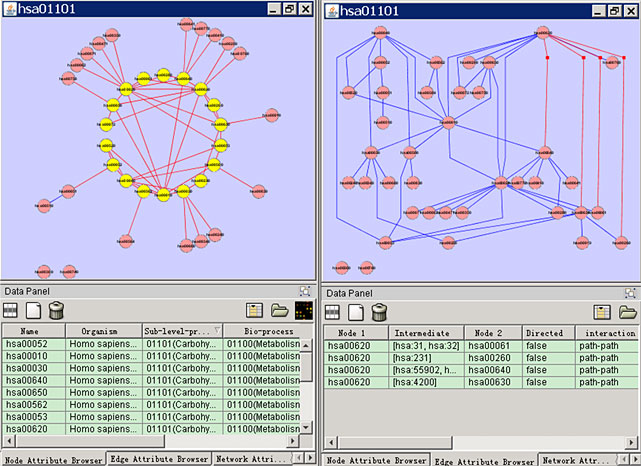
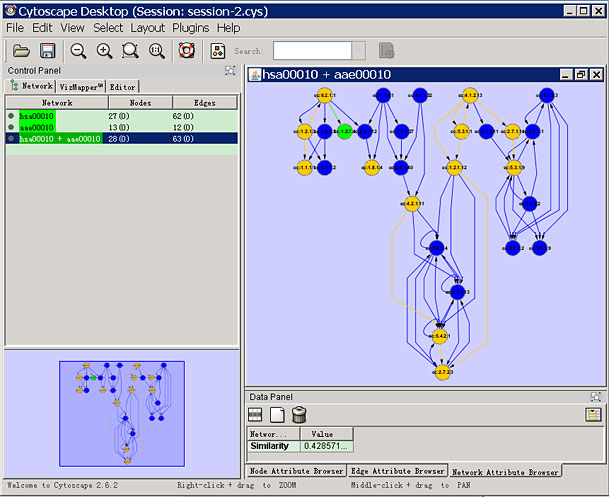
License
MetaGen is free under the GNU LGPL license. For the non-academic users, please check the license agreement of KEGG before using it.
Acknowledgement
We thank KEGG technical support
for the use of KEGG API.
We also thank the tech team of Cytoscape for the help on the plug-in
development. MetAtlas is funded by National Natural Science
Foundation of China (6077 3021 and 6060 3054).
Contact
| Tingting Zhou: | grace dot tingting dot zhou at gmail dot com |
| Samuel Kin Fung Yung: | cskfyung at comp dot polyu dot edu dot hk |