Rationale
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a comprehensive knowledge repository and popularly regarded as one of the main resources for modeling metabolic networks. KEGG manages the manually curated pathway maps in the KEGG/PATHWAY database, organizes these pathways using the functional hierarchies in the KEGG/BRITE database, and provides a graphic interface for the navigation of these pathway maps through KEGG/Atlas. Pathway maps in KEGG are represented as static or semi-static graphs, which is fixed and typically not accessible by computer programs. To address this problem, a handful of tools have been developed recently, including PaVESy, VisANT, KEGGspider, MEGU, KGML-ED, MetaViz and the most recent, KEGGgraph.
These tools facilitate the navigation of KEGG pathway maps but they are purposely designed for the visualization and editing. They display all the details for the pathway graphs. For those who only need metabolic networks in the context of enzyme or others, it is not a happy work for the researchers to spend long time to figure out if these tools provided the filters to exclude the non-enzymatic elements or not, or how to configure the filters in the right way.
In this case, we develop MetaGen, which condenses the metabolic network modeling into one line of command. Users only need to input three parameters to specialize the type, scope and metabolic objects for the modeling, and then MetaGen will take care of the remainder for you in batch processing.
Technology
MetaGen retrieves the metabolic data from the KEGG ftp site using the KEGG API and store data locally using a relational database server. MetaGen creates graph objects using JGraphT. Currently Pajek is the only supported network output format. More formats will be included soon.
MetaGen is developed with Eclipse and Java Development Kit (JDK) 6.0, and requires JDK 5.0 or above to run the system. The use of Spring and Maven makes MetaGen highly extendable and readable. Using the standard SQL syntax, MetaGen ships pre-configured with the MySQL database. But one can easily switch to the database they are comfortable administering by updating the database connection property file.
Methods
MetaGen is designed to model the given metabolic objects - the biological process (the entire metabolism), sub-level processes and metabolic pathways - into enzyme graphs and pathway graphs in batch processing. Firstly, according to the pre-set arguments, MetaGen visits KEGG to retrieve the data for use. Then MetaGen writes the data into the local database as soon as it models the metabolic objects into graphs. Only at the first time does MetaGen visit KEGG and retrieve data on a large scale. After that, MetaGen use the local data directly. In the local database, data have the time limit. Different section can have different pre-set expirations. From the second modeling of the same object, MetaGen checks the local data before it models the metabolic networks. If the local data hasn't expired, MetaGen uses the local data for the modeling directly, otherwise it visits KEGG and update the expired data first. The figure below shows the overall work scheme.
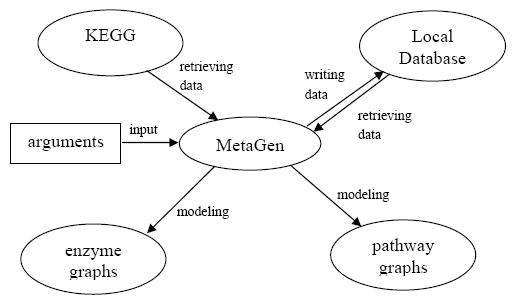
On the pathway level, MetaGen models metabolic pathways into enzyme graphs directly. It makes use of KEGG web service to retrieve enzymes and enzyme relations in that pathway and further assembles them into the enzyme graph model.
On the sub-process level or the bio-process level, there are three steps MetaGen takes to model the metabolic objects into enzyme graphs. First MetaGen looks up the KO hierarchy in order to know all the pathways belonging to the sub-level process or the entire metabolism; then MetaGen will model all the pathways into enzyme graphs one after another; finally MetaGen unites all these enzyme graphs into the larger one that is right the enzyme graph for the sub-level process or the entire metabolism.
Similarly is the process for MetaGen to model sub-level processes or the entire metabolism into pathway graphs. First MetaGen looks up the KO hierarchy to know all the involved pathways; then MetaGen will scan pathways one after another to retrieve all the linking pathway pairs. Having filtered out pathways and pathway links which are not in the metabolic object under investigation, MetaGen assembles the linking pathway pairs into pathway graphs as request.
Usage
MetaGen can work in two ways, for instance,
> java -jar metagen.jar e one hsa01101
or
> java -jar metagen.jar [command_file]
In case only several metabolic networks need to be modeled, you may consider the first way. In this way you need to input the three arguments one by one. None could be skipped. The first argument, 'e' or 'p', specifies the graph type for modeling to enzyme graph or pathway graph respectively. The second argument, 'one' or 'all', specifies the scope of the modeling. If 'one' is set, MetaGen just generates the one graph for the given metabolic object, and would not involve the graphs whose KO number is under this one's in the KO hierarchy. If 'all' is set, the latter. The third arguments is to give the organism-specific metabolic object to model. The 3- or 4-letter prefix is to address the organism, whose full name can be looked up at KEGG Organism. The 5-digit string is the KO number of the metabolic object. For example, '01100' refers to metabolism; '01101'~'01111' refers to the 11 sub-level processes; '00010' and so on refers to the metabolic pathways. This number of each metabolic objects can be looked up at KEGG ORTHOLOGY of pathway maps.
In case a bunch of graphs, especially the enzyme graphs on pathways, need to be modeled, you may like to consider generating a flat file as "command_file", which is the second way mentioned above. The content of "command_file" is just the collection of argument lines, for example,
e one hsa00010
p one hsa01101
e all hsa01100
Please note that there are no pathway graphs on pathway level to
be modeled. That is to say, 'p one hsa00010' doesn't make sense.
For more details, please check README.txt inside the package for downloading.
Acknowledgement
We thank KEGG technical support for the usage of KEGG API. Special thanks are also addressed to Prof Amos Bairoch for the helpful discussion. MetaGen is funded by National Natural Science Foundation of China (6077 3021 and 6060 3054).Contact
| Tingting Zhou: | grace dot tingting dot zhou at gmail dot com |
| Samuel Kin Fung Yung: | cskfyung at comp dot polyu dot edu dot hk |